7 Model evaluation: Part 2
When you get to class
- Open today’s Rmd file linked on the day-to-day schedule. Name and save the file.
- Knit the Rmd. Notice that it looks different because I added a theme at the top. This is totally optional but fun! To see other themes, visit here. These are just some options for customizing the appearance of your work.
Announcements
- Homework 2 and the next checkpoint are due on Tuesday (in 5 days).
- Quiz 1 is in two weeks. Please see details on page 7 of the syllabus. I recommend that you review each activity and homework, trying to answer the questions without first looking at your notes.
7.1 Getting started
WHERE ARE WE?
We’ve been building and, subsequently, evaluating statistical models. To this end, there are 3 important questions to ask:
- Is it wrong?
- Is it fair?
- Is it strong?
EXAMPLE
The high_peaks data includes information on hiking trails in the 46 “high peaks” in the Adirondack mountains of northern New York state.4

Our goal will be to understand the variability in the time in hours that it takes to complete each hike. In doing so, we’ll separately consider four possible predictors: a hike’s vertical ascent (feet), highest elevation (feet), length (miles), and difficulty rating (easy / moderate / difficult).
# Load useful packages and data
library(ggplot2)
library(dplyr)
peaks <- read.csv("https://www.macalester.edu/~ajohns24/data/high_peaks.csv")Construct a separate model of time by each predictor:
# Construct a model of time vs length
model_1 <- lm(time ~ length, data = peaks)
# Construct a model of time vs rating
model_2 <- lm(time ~ rating, data = peaks)
# Construct a model of time vs elevation
model_3 <- lm(time ~ elevation, data = peaks)
# Construct a model of time vs ascent
model_4 <- lm(time ~ ascent, data = peaks)To counter these numerical summaries, obtain a graphical summary of each relationship. Which relationship appears to be the strongest? Or, which variable seems to be the best predictor of the time required to complete the hike?
# Construct a plot of time alone# Construct a plot of time vs length# Construct a plot of time vs elevation# Construct a plot of time vs ascent# Construct a plot of time vs rating
Measuring model quality
R2 provides one rigorous measure of the strength of our model. The following plots illustrate the range of possible R2 values:
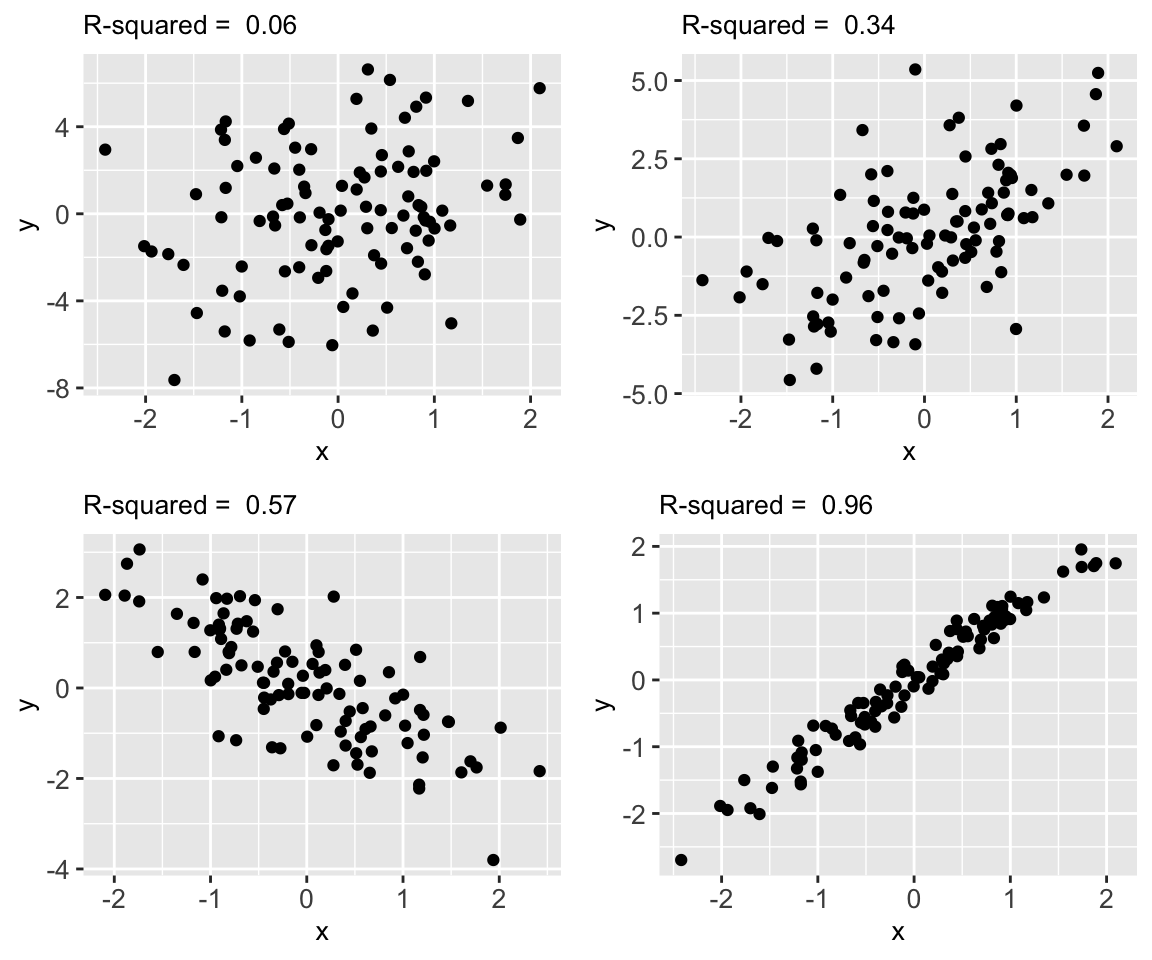
Interpreting R2
R2 measures the proportion of the variability in the observed response values that’s explained by the model. Thus 1 - R2 is the proportion of the variability in the observed response values that’s left UNexplained by the model.
\(0 \le R^2 \le 1\)
The closer R2 is to 1, the better the model.
7.2 Exercises
GOALS
Explore R-squared while also reviewing some other modeling concepts.
Warm-up: Data drill
Complete the following tasks usingdplyrverbs (filter,summarize,select,mutate,group_by) or using functions likenrow(),head(), etc.# How many hikes are in the data set? # Calculate the average elevation # Calculate the average elevation for each rating # Define a new variable, length_km, that records the length of a hike in km # NOTE: 1 mile is roughly 1.6 kilometers # Calculate the average hiking time # Calculate the average hiking time for hikes that are more than 14 miles long
Model review: model 1
Re-examine the below plot and model of the relationship between hikingtime(in hours) and hikelength(in miles). Write down the model formula, interpret the two model coefficients, and use this model to predict the time it takes to complete a 16-mile hike.# Plot the relationship ggplot(peaks, aes(y = time, x = length)) + geom_point() + geom_smooth(method = "lm", se = FALSE)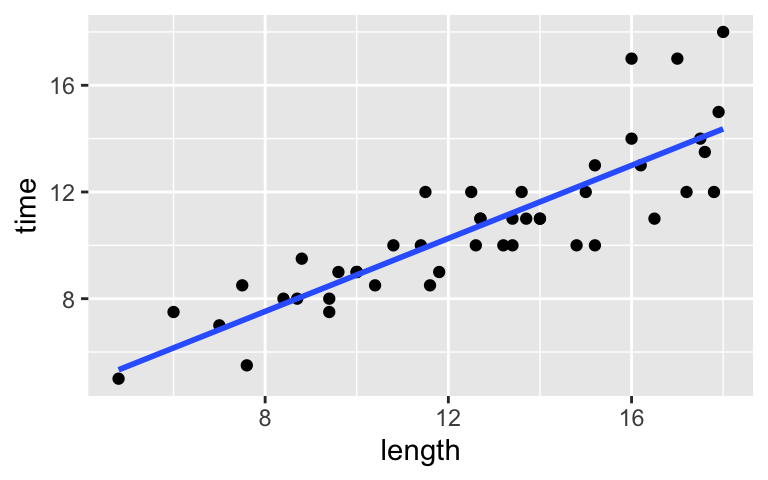
# Summarize the model summary(model_1) ## ## Call: ## lm(formula = time ~ length, data = peaks) ## ## Residuals: ## Min 1Q Median 3Q Max ## -2.4491 -0.6687 -0.0122 0.5590 4.0034 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 2.04817 0.80371 2.548 0.0144 * ## length 0.68427 0.06162 11.105 2.39e-14 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.449 on 44 degrees of freedom ## Multiple R-squared: 0.737, Adjusted R-squared: 0.7311 ## F-statistic: 123.3 on 1 and 44 DF, p-value: 2.39e-14
- Calculating R-squared: model_1
The above scatterplot suggests a relatively strong association between hike time and length. Using R-squared, let’s get a more precise numerical summary of this strength.Upon reviewing
summary(model_1)from the previous exercise, confirm that the relationship betweentimeandlengthhas an R-squared value of 0.737. NOTE: Focus onMultiple R-squaredand ignoreAdjusted R-squared(which we’ll discuss later in the semester).Confirm that we can also obtain R-squared using
summary(model_1)$r.squared.Interpret the R-squared value.
Model review: model 2
Re-examine the below plot and model of the relationship between hikingtime(in hours) and hikerating. Write down the model formula, interpret the three model coefficients, and use this model to predict the time it takes to complete an easy hike.# Plot the relationship ggplot(peaks, aes(y = time, x = rating)) + geom_boxplot()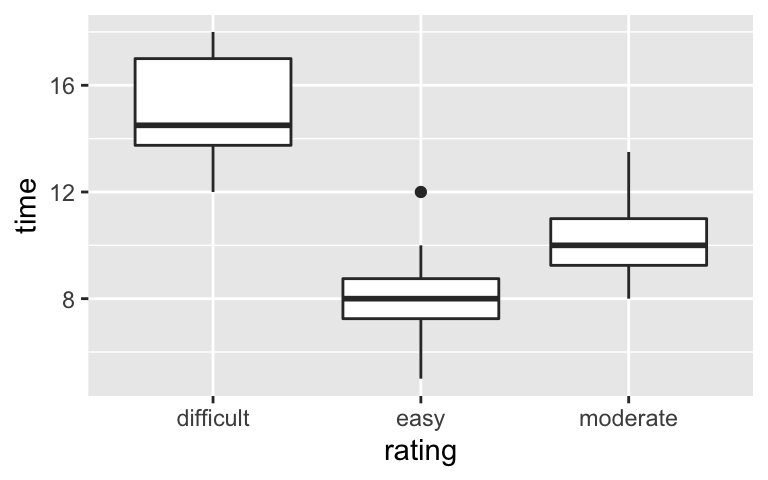
# Summarize the model summary(model_2) ## ## Call: ## lm(formula = time ~ rating, data = peaks) ## ## Residuals: ## Min 1Q Median 3Q Max ## -3.0000 -1.0000 -0.2222 0.8889 4.0000 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 15.0000 0.5947 25.222 < 2e-16 *** ## ratingeasy -7.0000 0.7816 -8.956 2.20e-11 *** ## ratingmoderate -4.5556 0.6771 -6.728 3.19e-08 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.682 on 43 degrees of freedom ## Multiple R-squared: 0.6538, Adjusted R-squared: 0.6377 ## F-statistic: 40.6 on 2 and 43 DF, p-value: 1.246e-10
- Calculating R-squared: model_2
Calculate and interpret the R-squared value formodel_2.
Comparing R-squared values
Finally, check out the R-squared values for each model:summary(model_1)$r.squared ## [1] 0.7370358 summary(model_2)$r.squared ## [1] 0.6538002 summary(model_3)$r.squared ## [1] 0.0002649341 summary(model_4)$r.squared ## [1] 0.219844- Which is the strongest model of hiking
time? Similarly, what is the strongest predictor of hiking time? - Which is the weakest model? Similarly, what is the weakest predictor of hiking time?
- Which is the strongest model of hiking
OPTIONAL digging deeper: part 1
Now that we’ve used R-squared, you might wonder how it’s calculated. If so, and especially if you might take more STAT classes, please try these optional exercises. But if you already have enough to chew on, please start working with your group on Homework 2.To learn about how R-squared is calculated, you’ll use some code that’s specialized to this task. We won’t use it more broadly, so don’t worry about understanding each detail.
Consider
model_1which modelstimebylength. For each hike, we can combine the observed completion time with the model residuals and predictions:# model_1 contains the predictions & residuals for each hike model_1$fitted.values model_1$residuals# Combine the observed values, predictions, residuals for each hike model_1_info <- peaks %>% select(time) %>% mutate(predicted = model_1$fitted.values, residual = model_1$residuals)Check out the results. Notice that the observed
timevalues, the predictedtimevalues, and the residuals all vary from hike to hike:head(model_1_info)Calculate the variance of the observed
timevalues,predictedvalues, andresidualvalues formodel_3.Repeat this process for
model_4, the model oftimebyascent. That is, calculate the variances of the observedtimevalues as well as the variances of the predictions and residuals calculated frommodel_1.
OPTIONAL digging deeper: part 2
Your work formodel_1andmodel_4is summarized below, along with additional calculations frommodel_3.Model Var(observed) Var(predicted) Var(residuals) R2 model_17.810 5.756 2.054 0.74 model_47.810 1.717 6.093 0.22 model_37.810 0.002 7.808 0.0003
- No matter the model, the variances of the observed
timevalues are always the same (since we’re using the same rawtimevalues in each model). Further the variances of the observedtimevalues, predictions, and residuals always have the same mathematical relationship. What is it?
- In general, which models have the most variable predictions and the least variable residuals – the models with higher or lower R-squared?
- No matter the model, how is R-squared calculated from the variance of the observed values and the variance of their predictions? Congrats! You’ve just established the formula for R2.
- Putting parts c and b together, explain why part b makes sense. That is, why is it desirable for the variability in the predictions to be similar to that in the observed y values? Or, why is it desirable to have small variance in the residuals?
- No matter the model, the variances of the observed
7.3 OPTIONAL – a formula for R-squared
We can partition the variance of the observed response values into the variability that’s explained by the model (the variance of the predictions) and the variability that’s left unexplained by the model (the variance of the residuals):
\[\text{Var(observed) = Var(predicted) + Var(residuals)}\]
“Good” models have residuals that don’t deviate far from 0. So the smaller the variance in the residuals (thus larger the variance in the predictions), the better the model! In pictures:

Putting this together, the R-squared compares Var(predicted) to Var(response):
\[R^2 = \frac{\text{variance of predicted values}}{\text{variance of observed response values}} = 1 - \frac{\text{variance of residuals}}{\text{variance of observed response values}}\]
Finally, IF you like mathematical formulas: Suppose we have a sample of \(n\) response values \((y_1,...,y_n)\) that have an average (mean) of \(\overline{y}\). Let \(\hat{y}_i\) be the prediction of \(y_i\) calculated from the model and \(y_i - \hat{y}_i\) be the corresponding residual. Then
\[ R^2 = \frac{\sum_{i=1}^n(\hat{y}_i - \overline{y})^2}{\sum_{i=1}^n(y_i - \overline{y})^2} = 1 - \frac{\sum_{i=1}^n(y_i - \hat{y}_i)^2}{\sum_{i=1}^n(y_i - \overline{y})^2} \]
7.4 Solutions
Warm-up: Data drill
# How many hikes are in the data set? nrow(peaks) ## [1] 46 # Calculate the average elevation peaks %>% summarize(mean(elevation)) ## mean(elevation) ## 1 4405.283 # Calculate the average elevation for each rating peaks %>% group_by(rating) %>% summarize(mean(elevation)) ## # A tibble: 3 × 2 ## rating `mean(elevation)` ## <chr> <dbl> ## 1 difficult 4527. ## 2 easy 4273 ## 3 moderate 4423. # Define a new variable, length_km, that records the length of a hike in km # NOTE: 1 mile is roughly 1.6 kilometers peaks %>% mutate(length_km = length * 1.6) %>% head() ## peak elevation difficulty ascent length time rating length_km ## 1 Mt. Marcy 5344 5 3166 14.8 10.0 moderate 23.68 ## 2 Algonquin Peak 5114 5 2936 9.6 9.0 moderate 15.36 ## 3 Mt. Haystack 4960 7 3570 17.8 12.0 difficult 28.48 ## 4 Mt. Skylight 4926 7 4265 17.9 15.0 difficult 28.64 ## 5 Whiteface Mtn. 4867 4 2535 10.4 8.5 easy 16.64 ## 6 Dix Mtn. 4857 5 2800 13.2 10.0 moderate 21.12 # Calculate the average hiking time peaks %>% summarize(mean(time)) ## mean(time) ## 1 10.65217 # Calculate the average hiking time for hikes that are more than 14 miles long peaks %>% filter(length > 14) %>% summarize(mean(time)) ## mean(time) ## 1 13.43333
- Model review: model 1
- time = 2.04817 + 0.68427length
- It doesn’t make sense to really interpret the intercept, the expected time to complete a “0-mile hike”.
- For each additional mile, we expect the hike to take 0.68 more hours (~41 more minutes) to complete.
- We expect it to take around 13 hours.
2.04817 + 0.68427*16 ## [1] 12.99649 - time = 2.04817 + 0.68427length
Calculating R-squared: model_1
summary(model_1)$r.squared ## [1] 0.7370358Hike length explains roughly 74% of the variability in completion times from hike to hike.
- Model review: model 2
- time = 15 - 7ratingeasy - 4.5556ratingmoderate
- We expect a difficult hike to take around 15 hours to complete.
- We expect an easy hike to take 7 hours less to complete than a difficult hike.
- We expect a moderate hike to take 4.5556 hours less to complete than a difficult hike.
- We expect an easy hike to take 8 hours (15 - 7).
- Calculating R-squared: model_2
\(R^2 = 40.6\). The difficulty of a hike explains roughly 40.6% of the variability in completion times from hike to hike.
- Comparing R-squared values
model_1. hike length is the strongest predictor.model_3. hike elevation is the weakest predictor.
OPTIONAL digging deeper: part 1
# Combine the observed values, predictions, residuals for each hike model_1_info <- peaks %>% select(time) %>% mutate(predicted = model_1$fitted.values, residual = model_1$residuals) head(model_1_info) ## time predicted residual ## 1 10.0 12.175427 -2.1754272 ## 2 9.0 8.617203 0.3827973 ## 3 12.0 14.228249 -2.2282490 ## 4 15.0 14.296676 0.7033236 ## 5 8.5 9.164622 -0.6646219 ## 6 10.0 11.080589 -1.0805889 # a model_1_info %>% summarize(var(time), var(predicted), var(residual)) ## var(time) var(predicted) var(residual) ## 1 7.809662 5.756 2.053662 # b model_4_info <- peaks %>% select(time) %>% mutate(predicted = model_4$fitted.values, residual = model_4$residuals) model_4_info %>% summarize(var(time), var(predicted), var(residual)) ## var(time) var(predicted) var(residual) ## 1 7.809662 1.716908 6.092754
- OPTIONAL digging deeper: part 2
- var(observed values) = var(predictions) + var(residuals)
- models with higher R-squared
- var(predictions) / var(observed values)
- We want the predictions to be as similar to the observed values as possible, thus we want the variability in the predictions to be similar to that in the observed y values. Similarly, we want the predictions to vary as little from 0 as possible.
original data source =
HighPeaksdata in theStat2Datapackage; original image source = https://commons.wikimedia.org/wiki/File:New_York_state_geographic_map-en.svg↩︎